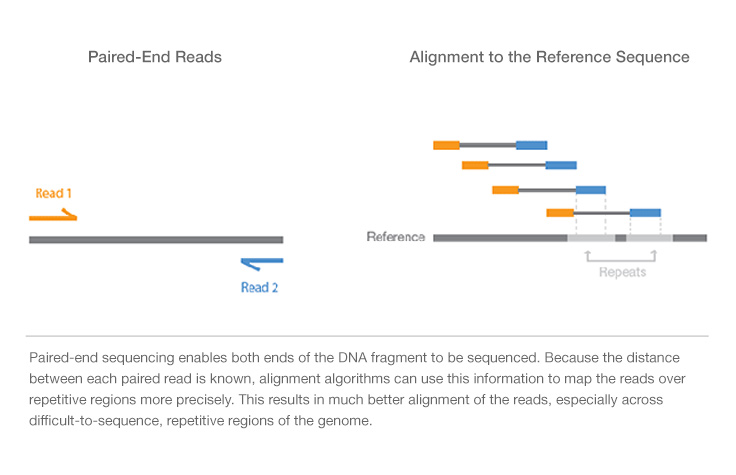
paired end sequencing vs mate pair
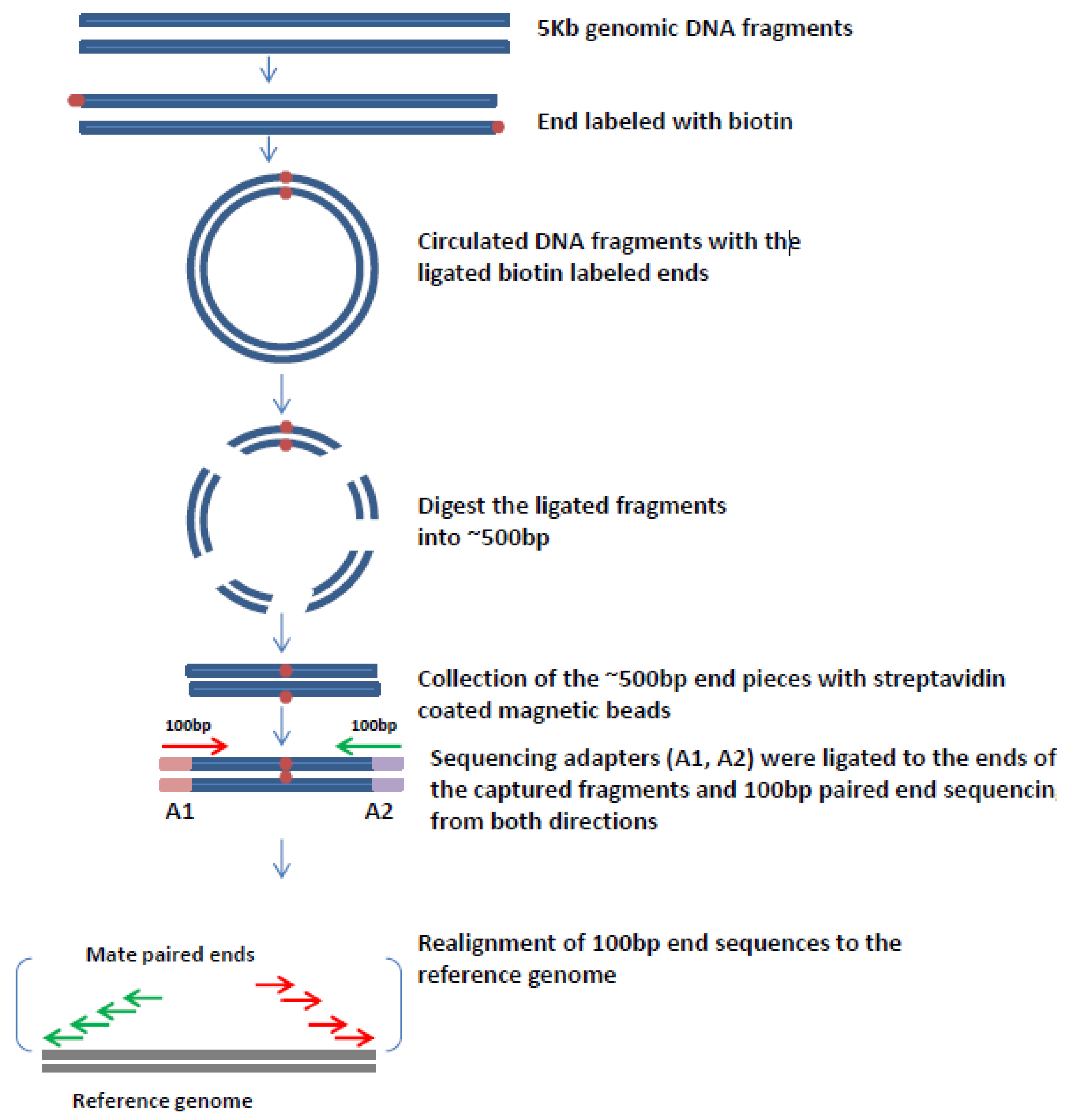
The figure shows the workflow for mate-pair library preparation for Illumina sequencing. Introduction to Mate Pair Sequencing.

Ion Torrent Mate Paired Library Preparation Protocol And Sequencing Download Scientific Diagram
Mate-pair is a specific type of library.

. Mate-pair sequencing은 paired-end read를 긴 insert와 함께 생성하는 방법. What are paired end reads Illumina. Usually mate-pair library are used to identify structural.
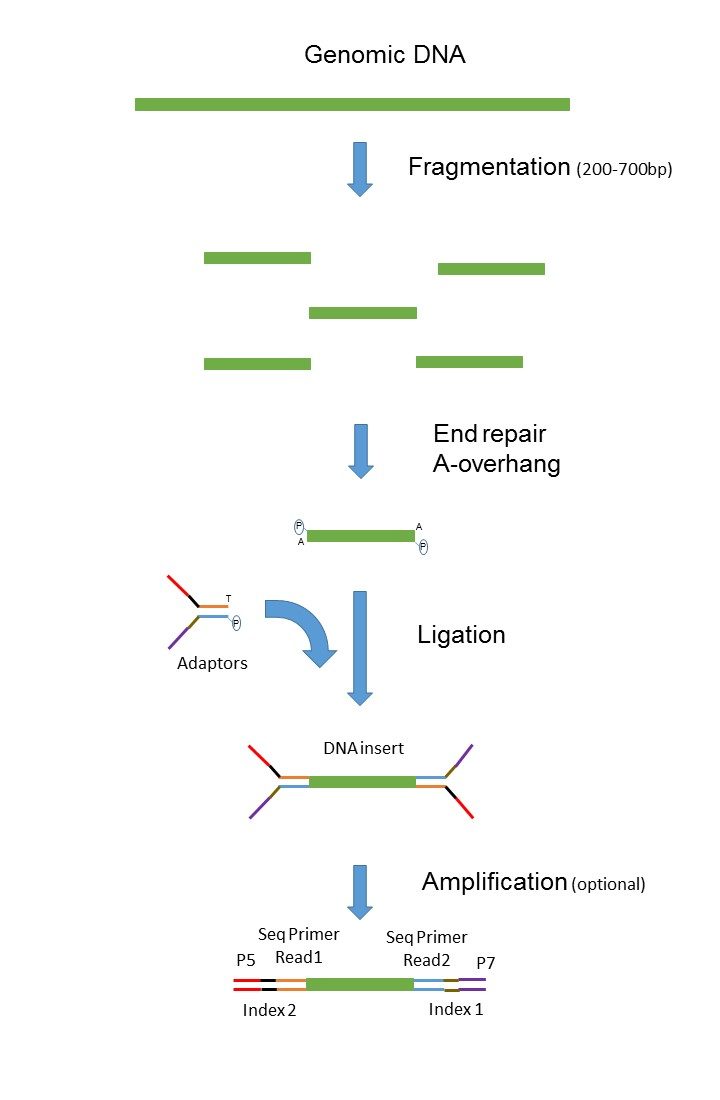
The differences between PE and MP reads include. Library preparation protocols -- In short PE protocols attach an adapter SP1 to the fwd end and another adapter SP2 to the reverse end. Mate pair sequencing enables generation of long-insert paired-end DNA libraries for de novo sequencing structural variant detection and other applications.
후자가 mate-pair라고 불리는 것으로 둘의 차이는 insert 길이의 차이임. This can be done using either optical mapping or mate-pair sequencing. Requires the same amount of DNA as single-read genomic DNA or cDNA sequencing.
The second sequencing step targets SP2 to generate the reverse read. The first sequencing step is started by targeting SP1 to generate the forward read. When a DNA fragment is shorter than two times the read length the paired reads overlap and can be merged into a longer read.
Can be used for. Standard libraries depends upon your application. Also these libraries have insert sizes much longer than the paired end.
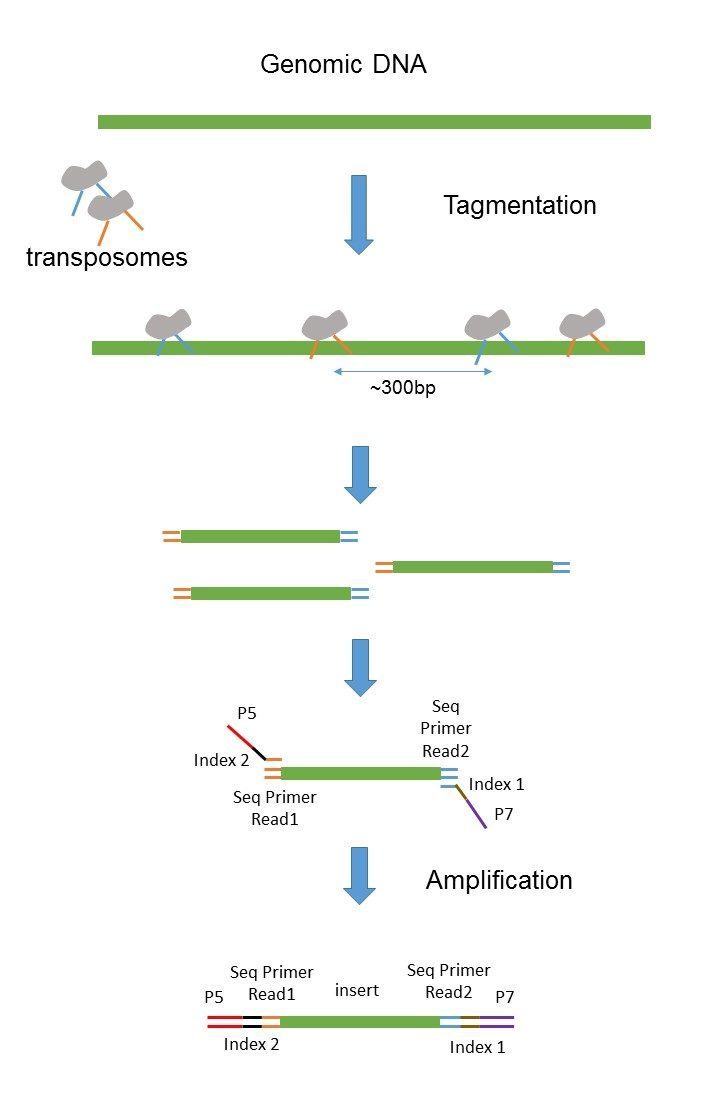
Paired end や mate pair という用語はどのようにライブラリが作られたかどうやってシーケンスされたかを示します. Mate pair allows you to have your pairs be much farther apart which can be more. Since the beginning of 2013 this preparation has been based on Nextera.
Bases 1-75 forward direction and bases 225-300 reverse direction of the fragment. 50 base-pair paired-end reads span a longer region of the transcript. Combining data generated from mate pair library sequencing with that from short-insert paired-end reads provides a powerful combination of read lengths for maximal.
The preparation of mate pair libraries is designed to allow classical paired-end sequencing of both ends of a fragment with an original size of several kilobases. Paired-End Sequencing - Acheving maximum coverage across the genome Illumina Mate Pair Library Sequencing - Characterization genome variation Illumina 플라스미드에 클로닝하여 만든. Simple workflow allows generation of unique ranges of insert sizes.
For example if you have a 300bp contiguous fragment the machine will sequence eg. Show activity on this post. 50 bp of sequence in a mate pair can be more useful for read mapping than an extra 50 bp in the read itself.
They are not two different methods. If you dont. Does not require methylation of DNA or restriction digestion.
Paired end sequencing reffers to sequrncing of fragments from both ends this is in contrast to single end sequemcing where sequencing is done from one end. Fortunately Illumina offers paired-end PE reads which are sequences at the two ends of DNA fragments. The decision to use mate-pair vs.
Illumina에서 이야기하는 mate pair library는 일종의 jumping library라고 하는 것이 기술적으로 더 정확할 수 있겠다. Paired-end sequencing에서 두 종류의 read short-insert paired-end reads SIPERs long-insert paired-end reads LIPERs를 만들 수 있음. In paired-end sequencing the library preparation yields a set of fragments and the machine sequences each fragment from both ends.
Paired-end is a type of sequencing. In fact mate-pair libraries require paired-end sequencing. Learn about the difference between Paired-End and Single-Run sequencing and why the former creates more precise alignments than the latter especiall.
Broad Range of Applications. Each read represents one end of a 200-300 base-pair RNA fragment compared to a 100 base-pair read which only gives you information about 100 bases. Paired-end tags PET sometimes Paired-End diTags or simply ditags are the short sequences at the 5 and 3 ends of a DNA fragment which are unique enough that they theoretically exist together only once in a genome therefore making the sequence of the DNA in between them available upon search if full-genome sequence data is available or upon further.
Another supposed advantage is that it leads to more accurate reads because if say Read 1 see picture below maps to two different regions of the genome Read 2 can be used to help determine which one of the two. All Illumina next-generation sequencing NGS systems are capable of paired-end. One of the advantages of paired end sequencing over single end is that it doubles the amount of data.
Paired-end sequencing facilitates detection of genomic rearrangements and repetitive sequence elements as well as gene fusions and novel transcripts. Mate pair sequencing involves generating long-insert paired-end DNA libraries useful for a number of sequencing applications including. 例えばゲノム DNA を.
Since paired-end reads are more likely to align to a reference the quality of the entire data set improves. In the range of kb ie 1kb 2kb or longer in some cases.
Why Mate Paired End Paired End And Single End Reads Library To Be Combined For Assembling
Paired End Sequencing France Genomique
Paired End Sequencing France Genomique

What Is Mate Pair Sequencing For
Viruses Free Full Text Mate Pair Sequencing As A Powerful Clinical Tool For The Characterization Of Cancers With A Dna Viral Etiology Html

6 Pairwise Strand Of Paired End And Mate Pair Read Libraries Download Scientific Diagram

Schematic Representation Of Mate Pair Library Construction By Coupling Download Scientific Diagram

Pdf Data Processing Of Nextera Mate Pair Reads On Illumina Sequencing Platforms Semantic Scholar

Mate Pair Made Possible Enseqlopedia
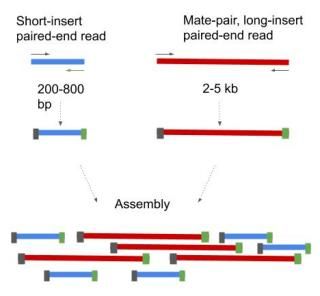
Introduction To Genome Assembly Tutorial

What Is Mate Pair Sequencing For
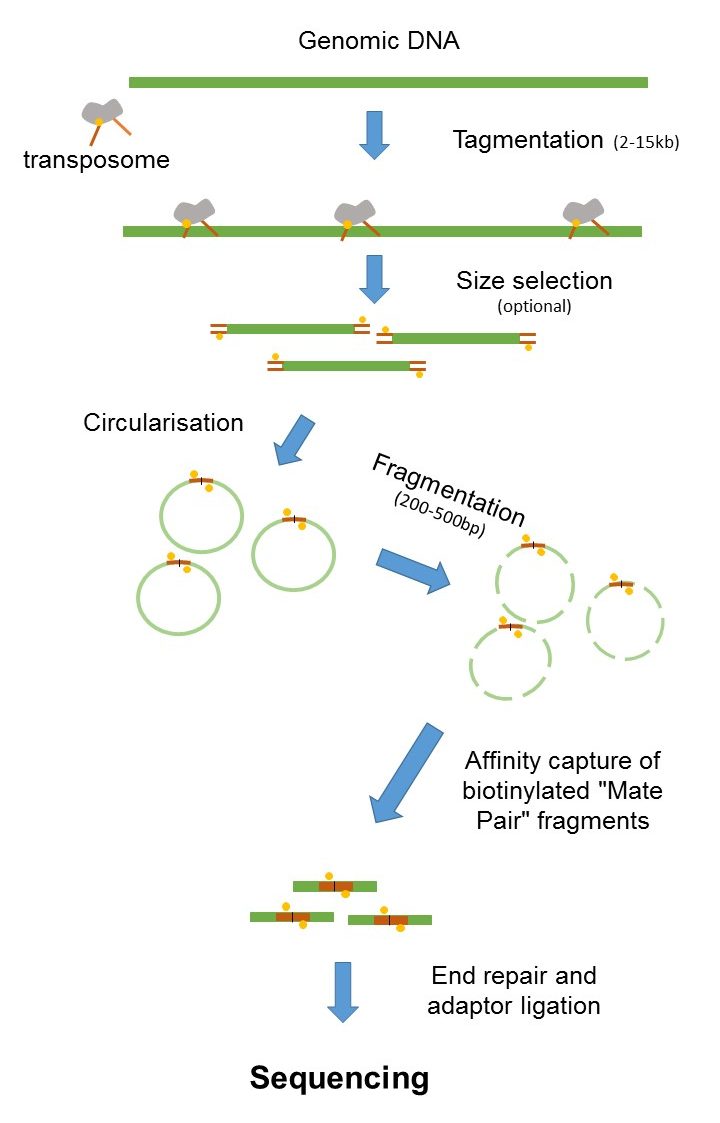
Mate Pair Sequencing France Genomique

Beyond The Linear Genome Paired End Sequencing As A Biophysical Tool Trends In Cell Biology

Construction Of Mate Pair Libraries Download Scientific Diagram
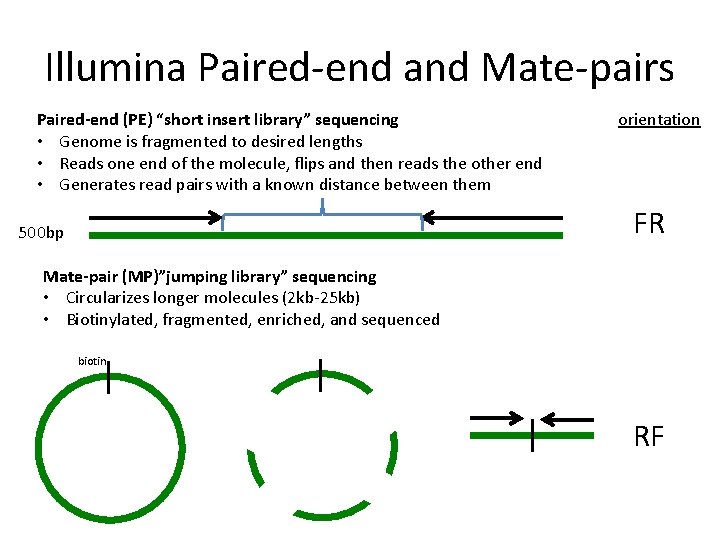
De Novo Genome Assembly Using Next Generation Sequence

Use Of Pairwise Linkage Information For Scaffolding A Paired End Download Scientific Diagram